Abstract
Every geneticist interested in genome evolution will have to face the analysis and comparison
of strains of plasmids and viruses. For this purpose, several plasmids resources have been
developed very recently and propose either a comprehensive manually curated bacterial plasmid
list or a plasmid database which can be interrogated using sequence similarity programs.
To our knowledge, bioinformatics tools providing geneticists with deeper plasmid analyses
such as extensive gene synteny are still missing at this time. Widely used existing prokaryotic
synteny web services are mostly geared towards cellular genomes and provide at most limited
plasmid analysis capabilities. Here we present the Natural Plasmid DataBase (WASPS)
which is developed in a plasmid-centric fashion and proposed to the researcher as a straightforward
web service. The novelty of WASPS consists in pre-calculated orthologous clusters covering all
plasmid-encoded proteins available in the database. The ‘fully connected’ clustering topology
allows the near-instantaneous generation of completely resolved synteny maps. The relevant
features of WASPS include: 1) text-based search in plasmid definitions and accession numbers;
2) protein and DNA-based similarity searches with user-provided sequences; 3) synteny mapping
using WASPS plasmids as query; 4) synteny mapping using a user-provided annotated file in Genbank
format and 5) synteny mapping using unannotated raw or Fasta DNA file. Features 3 to 5 involving
the display of gene synteny maps benefit from technologies provided by latest Web standards.
Extensive use of the HTML5 canvas allows for a highly responsive visualization without server
recalculation. Wide genomic areas can be explored by panning and zooming directly within the
client browser. Mapping data can be exported either as .PNG bitmaps or under .SVG vectorial form.
The plasmid dataset originates from the NCBI plasmid repository and is converted into a compact
database locally on our server. The database compilation is optimized and fully automated to
allow frequent updates and ensure exhaustiveness of the analyses. It comprises all natural
bacterial, archaeal and eukaryal plasmids.
General description. The WASPS Natural Plasmid Database is composed of three independent components: the WASPS Database,
the WASPS Webtool and the WASPS Updater.
- The WASPS Database is a free standing entity designed to
be accessible locally via server applications
and scripts or remotely via specifically designed web services.
- The WASPS Webtool offers public access to the WASPS Database.
Remote users can perform a variety of
queries and retrieve several levels of information relative to a particular plasmid or investigate
the evolutionary relationship between different plasmids in the database using gene synteny. Similarly, user-provided
annotated or unannotated plasmid sequences can be analyzed as well.
- The WASPS Updater regenerates the whole WASPS Database at fixed intervals
to ensure maximal exhaustiveness of all plasmid analyses.
Synteny
- Etymology
Synteny = on the same ribbon
Greek: σύν (syn) = 'along with' and ταινία (tainiā) = 'band'
- Origin
The term 'synteny' was introduced by John H. Renwick at the 4th Int. Congress of Human Genetics in 1971.
“Synteny (or syntenic) refers to gene loci on the same chromosome whether or not they are genetically linked by classic linkage analysis.” (Renwick, 1971)
- Debate
Some considered that the usage of the term synteny was incorrect. “Synteny refers [incorrectly] to gene loci in different organisms located on a chromosomal
region of common evolutionary ancestry.” (Passarge, Bernhard & Farber, Nat. Gen. Corresp. 1999)
- Present concept of 'shared synteny'
The concepts of 'synteny' or 'shared synteny' are now commonly accepted and no longer controversial. Synteny constitutes the
most reliable criteria for establishing the orthology of genomic regions in different species and
reflects important functional relationships between genes.
- WASPS synteny maps
WASPS Webtool is designed to display evolutionary relationships between natural plasmids and user-submitted DNA sequences.
Each line corresponds to a single plasmid where multiple arrows refer to gene open reading frames (Fig.1).
The orthology between genes on the same plasmid or on different plasmids is indicated by a common color/pattern combination.
Patterns have been introduced to increase the number of orthologs that can displayed at once to overcome the limitation of using color hues only.
Genes represented in white color are singletons: they don't share orthology reltionships with any other gene in the map.
WASPS is calculating protein similarity (and not DNA) to assess gene orthology relationships relying on the following programs:
UCLUST, BLAST and DIAMOND depending on the option chosen.
WASPS orthologous clusters are established at database creation time and colors/patterns combinations are assigned to each cluster at run time.
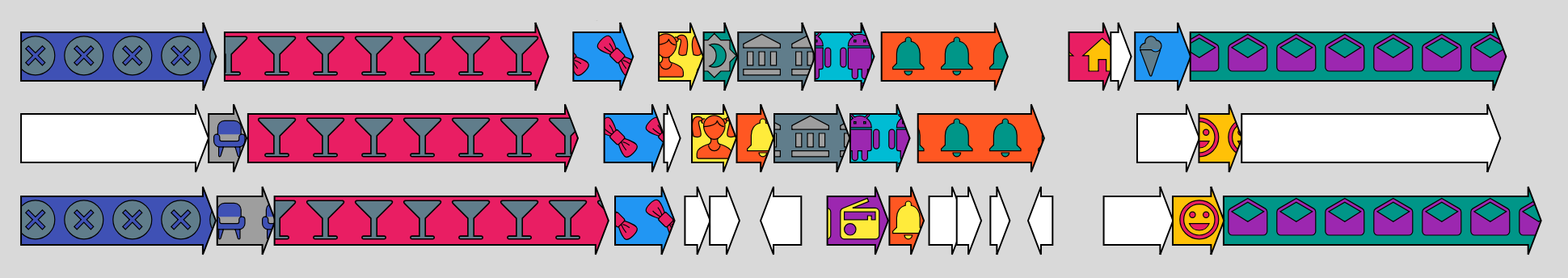 Figure 1. Plasmid synteny displayed in WASPS synteny maps.
Figure 1. Plasmid synteny displayed in WASPS synteny maps.
Gene orthology
An orthologous gene is a gene in different species that evolved from a common ancestor by speciation.
Orthologous genes retain the same function in the course of evolution.
1. WASPS Database
1.1 WASPS Database structure. Annotated plasmids entries (RefSeq) are retrieved form the NCBI FTP site in binary form and processed locally
on this server to generate the WASPS Database. The RefSeq plasmid records are updated on a regular bas on the
NCBI FTP site every 2 or 3 weeks. This process involves the addition of new entries which receive a two part
NCBI identifier (accession.version). Updates of pre-existing entries keep the same accession number and will
increment the version number by one unit. These ‘accession.version’ identifiers are used to univocally describe
both plasmid and gene entries. These identifiers are used to link the different data bins composing WASPS database.
The central part of the WASPS database consists of a single XML file containing a sequential list of plasmids exposing
their relevant fields. Each plasmid contains a gene list field to store relevant genetic data (Fig. 2). Each protein
in the WASPS database is therefore identified by double ‘accession.version’ under the format ‘gene_accession.version#plasmid_accession.version’.
Plasmid DNA sequences and protein sequences are stored in separate bins but intimately linked to the central XML using
the ‘accession.version’ identifiers. All fields and DNA or protein sequences extracted or parsed from the downloaded NCBI
GenBank and DNA Fasta files. Protein orthology relationships are determined using UCluster and injected where
appropriate in the XML file. The cluster centroids calculated with UClust are collected separately into an additional
bin. The text bins containing DNA, total proteins and centroid proteins sequences are then converted in binary format
in order to be efficiently queried by BlastN, BlastP, TBlastN or PsiBlast (Fig. 2).
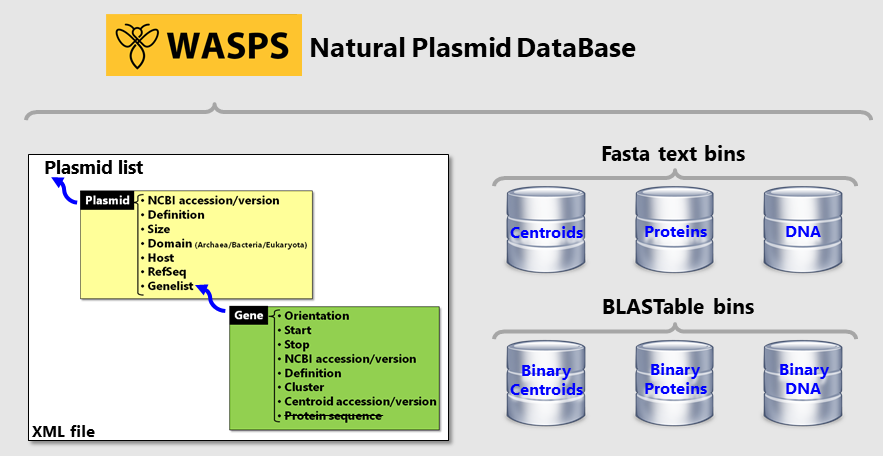
Figure 2. Structure of the WASPS Natural Plasmid Database.
2. WASPS Webtool
The WASPS Webtool provides the user with different ways to query the WASPS Database.
2.1 Text-based WASPS search. This is the simplest way to interrogate the WASPS Database. A user-provided text string will be matched against
four fields of the database: i) plasmid accession field; ii) plasmid definition field; iii) gene accession
field and iv) gene definition field. If the query is successful, the WASPS Webtool will provide relevant
‘accession.version’ numbers followed by the corresponding definition. Each hit will specify in addition
its plasmid or gene origin. ‘Accession.version’ identifiers obtained in this way can be used with other
WASPS Webtool queries/options. A domain-specific text search is also possible.
Text search results are connected to the plasmid synteny page via -> Synteny links to facilite further analyses.
2.2 Perform local plasmid synteny. A user-provided genetic or plasmidic ‘accession.version’ identifier will be matched against the database.
The webtool will extract the list of plasmids sharing orthologous relationships with the query gene or plasmid.
Plasmids from this list can be further selected to be displayed on the Synteny Map Interface.
Conservation
of protein encoding gene order or synteny is indicated by consistent icon & coloring. This map allows smooth
pan and zoom navigation using a three button wheel mouse. Synteny results can be exported in bitmap PNG
or vector SVG formats.
2.3 Perform external plasmid synteny (raw/Fasta). A user-provided plasmid sequence in raw or Fasta format (therefore unannotated)
will be scanned for open
reading frames (ORFs) in the six possible frames. These ORFs are translated into protein and matched
against the centroid database. Hits obtained below the preselected threshold E-value will be assigned
a cluster number and colorized accordingly. The search mode of related
plasmids is user-selected according to two
rules: i) display the primary hits only or ii) display all cluster-related hits. Plasmids from this list
can be further selected to be displayed on the Synteny Map Interface. Conservation of protein encoding
gene order or synteny is indicated by consistent icon & coloring.
The map will therefore display the predicted ORF results for the submitted sequence in the 6 reading frames
followed by the related plasmids from the WASPS Database. This map allows smooth pan and zoom navigation using a
three button wheel mouse. Synteny results can be exported in bitmap PNG
or vector SVG formats. Translated open
reading frames can be exported as protein sequences in Fasta format. Two option are available: export the complete set
or translated ORFs or only the subset which was assigned a predicted function by the WASPS Webtool.
2.4 Perform external plasmid synteny (GenBank). Annotated plasmids in GenBank format can be submitted by the user for further analysis. Annotated
proteins sequences are
extracted and matches against the centroid database. Hits obtained below the preselected threshold E-value will
be assigned a cluster number and colorized accordingly. The search mode of related
plasmids is user-selected according to two rules:
i) display the primary hits only or ii) display all cluster-related hits. Plasmids from this list can be further selected
to be displayed on the Synteny Map Interface. Conservation of protein encoding gene order
or synteny is indicated by consistent
icon & coloring. The map will therefore display the query plasmid on the first line followed by the related plasmids from
the WASPS Database. This map allows smooth pan and zoom navigation using a three button wheel mouse. Synteny results can be exported
in bitmap PNG or vector SVG formats.
2.5 Blast a protein or DNA sequence against WASPS. The WASPS Database can be queried with user-provided single DNA sequences (with BlastN) or protein sequences
(BlastP, TBlastN & PsiBlast).
Sequences can be submitted in raw or Fasta format. The results page displays an enhanced list of hits with embedded NCBI links
and the relative sequence alignments.
BLAST search results are connected to the plasmid synteny page via -> Synteny links to facilite further analyses.
2.6 WASPS Dashboard. The WASPS dashboard display statistical information on the database in graphical form.
2.7 WASPS Help. This page.
2.8 WASPS links. To come.
3. WASPS Updater
The WASPS Updater regenerates the whole database at fixed intervals to ensure maximal exhaustiveness of all plasmid analyses.
The WASPS update is a complex process which has been largely optimized for execution speed and low CPU resource consumption (Fig. 3).
It is completely unsupervised and automated and occurs at the frequency of NCBI RefSeq plasmid updates. Each update process
will regenerate a new database from scratch due to the fact that protein orthology calculations cannot be produced reliably
using incremental database updates. Plasmid data originates from the National Center for
Biotechnology Information (NCBI) and
is retrieved from their public access FTP site. At this stage, only the
RefSeq plasmid releases are considered in the WASPS Database because they
constitute a non-redundant and well-annotated set of sequences. The XML file described above constitutes the core of the WASPS
database and is stored on disk and
deserialized to server cache memory to ensure fast response across sessions/users. Alternative binary formats
were tested for database disk storage but their deserialization ranked lower that the XML file in benchmark
tests for this particular implementation.
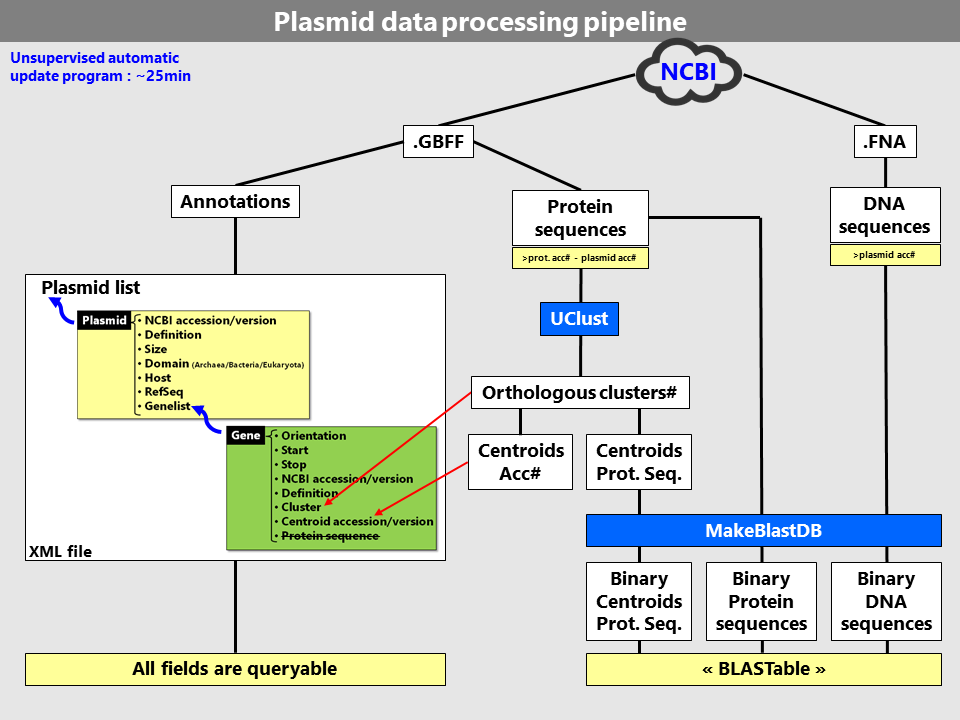
Figure 3. WASPS database generation pipeline.
4. Additional information
4.1 Input file formats. The
WASPS Webtool accepts user-submitted plasmid or protein data in three different formats:
-
Raw format.
The raw format is the simplest format which can be produced for proteins and DNA. It consists of a text string of amino
acids or nucleotides represented as single characters.
While carriage returns are accepted (and ignored) in this format, any other non-amino acid or non-DNA character with cause
submission failure (Fig. 4).

Figure 4. Raw sequence format.
-
Fasta format.
The Fasta format contains a comment line starting with the '>' character. The actual sequence string initiates at the second
line and is similar to the raw format (Fig. 5).
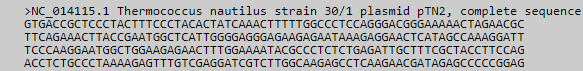
Figure 5. Fasta sequence format.
-
GenBank format.
The GenBank format contains a plethora of annotations, in addition to the complete sequence data at the end. GenBank obeys to
very strict formatting rules and conventions, it conveys all the available information relative to a specific sequence (Fig. 6).

Figure 6. GenBank sequence format.
4.2 UCLUST clustering algorithm.
UCLUST (Edgar, 2010) is a powerful clustering alorithm used by the WASPS
Database to precalculate protein orthologous clusters at database creation. UCLUST is also providing a centroid protein for each orthologous cluster.
The WASPS Webtool uses these centroids to quickly determine orthologous relationships at run time between
user provided sequences and the WASPS Database.
4.3 BLAST. Five similarity search options are available:
-
DIAMOND. This option, given a protein query, returns the most similar protein sequences from the WASPS protein database (using very fast DIAMOND).
-
BlastP. This option, given a protein query, returns the most similar protein sequences from the WASPS protein database (using classical NBCI BlastP).
-
BlastN. This option, given a DNA query, returns the most similar protein sequences from the WASPS DNA database.
-
TBlastN. This option compares a protein query against all six reading frames of the WASPS DNA database.
-
Psi-Blast. This option is used to find distant relatives of a protein in the WASPS protein database.
4.4 E-value. The lower the E-value, or the closer it is to zero, the more "significant" the match is.
4.5 Search mode. Currently two synteny search modes are provided by the WASPS Webtool.
-
Primary hits only. This option will only retrieve the plasmid primary hits which correspond to the best BlastP hit
for each protein against the WASPS centroid database
-
Extented cluster hits. This option will extent the best plasmid hits by adding all related plasmids using the WASPS
protein cluster database
4.6 Search algorithm.
To draw synteny maps using user-provided DNA sequences in raw, Fasta or GenBank formats,
the WASPS Webtool will extract corresponding protein sequences and match them against the WASPS centroid database bin.
They are used to assess the statistical significance of the sequence similarity using
the E-value.
All protein-protein similarity searches are performed locally on the WASPS server. Currently two similarity search algorithms are proposed by the WASPS Webtool.
-
BLAST. BLAST is one of the most widely used bioinformatics programs for sequence searching and is provided by the
National Center for Biotechnology Information (NCBI).
(Altschul et al, 1990)
-
DIAMOND. DIAMOND provides a fast and sensitive protein alignment algorithm (Buchfink et al, 2015).
Performance-wise, DIAMOND oupterforms in terms of speed and accuracy most competing similarity search programs.
The implementation of DIAMOND for orthologous protein assessment in WASPS is significantly faster in most cases but more variable than BLAST
depending on the server load.
4.7 Graphical export. Synteny maps generated by the WASPS Webtool can be exported in several formats for storage or further graphical elaboration.
-
PNG. Portable Network Graphics (PNG) is a raster-graphics non-patented file format that supports lossless data compression.
PNG files can be manipulated in pixel-oriented graphics programs such as Adobe Photoshop, Affinity Photo or Gimp.
For the WASPS Webtool, PNG export acts as a screenshot: only the visible parts of the synteny map will be exported.
-
SVG. Scalable Vector Graphics (SVG) is an XML-based vector image format for two-dimensional graphics.
The vectorial nature of this format allows substantial enlargement without resolution loss.
SVG files can be manipulated in vector-oriented graphics programs such as Adobe Illustrator, Affinity Designer or Inkscape.
4.8 Protein sequence export.
The synteny map produced with a user-submitted raw or Fasta plasmid sequence allows the export in Fasta format of i) all predicted translated ORFs or
ii) only the predicted translated ORFs that have been assigned a function by the WASPS Webtool.
4.9 Plasmid selection. Plasmids found by the WASPS Webtool are listed on the leftmost listbox.
Further processing to generate synteny maps require the presence of user-selected plasmids in the rightmost listbox.
A series of intuitive buttons allow plasmid movement between these two listboxes. A maximum of 50 different plasmids can be
visualized at once on the synteny maps, corresponding to the 50 topmost plasmids in the rigthmost listbox.
4.10 Synteny Map Interface navigation.
The Synteny Map Interface has been developed to allow a maximal user-interactivity with the genetics maps generated by the WASPS Webtool.
User interactivity is achieved my the means of an inexpensive 'three button wheel mouse', a standard equipment for most modern desktop
computers (Fig. 7). Equivalent gestures are available for laptop trackpads or touch screen devices and provided by the respective operating systems.

Figure 7. Three button wheel mouse.
2D synteny maps can be smoothly panned and zoomed directly in the web browser without requiring data transfer from or to the server.
This remarkable property has been developed by exploiting the latest D3 javascript libraries designed
to display Data Driven Documents.
-
Pan. The Synteny Map Interface can be panned by holding down the left mouse button.
-
Zoom. The Synteny Map Interface can be zoomed by holding by turning the mouse wheel.
-
Hovering. Context-sensistive information is available for each displayed gene in the synteny maps.
Mouse hovering on a specific gene will present its definition in a tootip (Fig. 8).
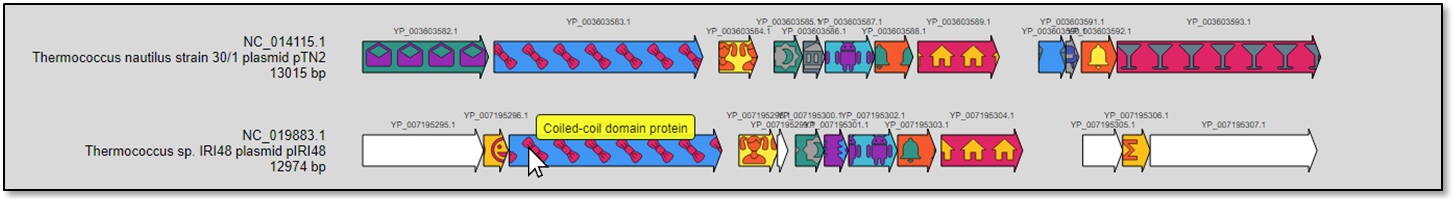 Figure 8. The hovering tooltip appears in yellow color.
Figure 8. The hovering tooltip appears in yellow color.
-
Context menu. Right clicking on a specific gene will open a context menu with four options (Fig. 9):
i)Info : protein gene accession, plasmid accession, protein definition and protein cluster.
ii)Single sequence : protein sequence of the highlighted gene in Fasta format.
iii)Multiple sequences : the whole protein cluster related to the highlighted gene.
iv)NCBI : link to the protein (Genbank format) at the NCBI.
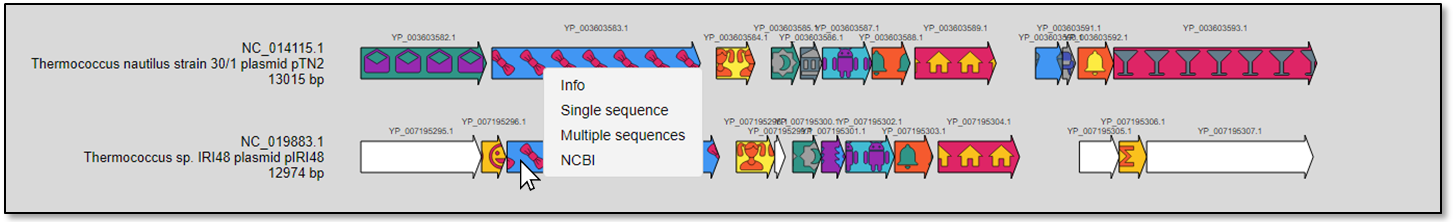 Figure 9. The context menu appears in white color.
Figure 9. The context menu appears in white color.
5. Further developments
- Granting REST
access to the database, allowing remote researchers to build their own web apps to exploit the WASPS Database.
- A comprehensive natural virus database built along the same model as WASPS.
6. References
-
Altschul S.F., Gish W., Miller W., Myers E.W. & Lipman D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215(3):403-10.
[PubMed]
-
Buchfink B., Xie C. & Huson DH. (2015) Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12(1):59.
[PubMed]
-
Edgar R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 1;26(19):2460.
[PubMed]
-
Passarge E., Horsthemke B. & Farber R.A. (1999) Incorrect use of the term synteny. Nat Genet. 23(4):387.
[PubMed]
-
Renwick J.H. (1971) The mapping of human chromosomes. Annu Rev Genet. 5:81-120.
[PubMed]