BAGET 2.0
Bacterial and Archaeal Gene Exploration Tool
BAGET Help
Abstract
Motivation: The retrieval of a single gene sequence and context from completely sequenced bacterial and archaeal genomes constitutes
an intimidating task for the wet bench biologist. Existing web-based genome browsers are either too complex for routine use or only
provide a subset of the available prokaryotic genomes.
Results: We have developed BAGET 2.0 (Bacterial and Archaeal Gene Exploration Tool), an updated web service granting access in just three mouse clicks to the sequence and synteny of any gene from completely sequenced bacteria and archaea. User-provided annotated genomes can be processed as well. BAGET 2.0 relies on a local database updated on a daily basis.
Availability and implementation: BAGET 2.0 befits all current browsers such as Chrome, Firefox, Edge, Opera and Safari. Internet Explorer 11 is supported as well.
Results: We have developed BAGET 2.0 (Bacterial and Archaeal Gene Exploration Tool), an updated web service granting access in just three mouse clicks to the sequence and synteny of any gene from completely sequenced bacteria and archaea. User-provided annotated genomes can be processed as well. BAGET 2.0 relies on a local database updated on a daily basis.
Availability and implementation: BAGET 2.0 befits all current browsers such as Chrome, Firefox, Edge, Opera and Safari. Internet Explorer 11 is supported as well.
Genome selection from the database
In order to ease the selection among the multiple (>50,000) DNA molecules accessible in the genome database, the user is
first invited to pick out the initial (A to Z) of the corresponding organism and then the complete genus-specied identifier.
When a given organisms harbors several independent replicons, they are named using the organism genus_species identifier followed
by the C1, C2.. suffices and ranked by decreasing size. Each replicon can be user-selected independently.
Genome database
The genome database accessible using BAGET comprises all the completely sequenced archaeal and bacterial genomes available from the NCBI
including the relative extrachromosomal elements and plasmids. The genome database is hosted locally on our servers to ensure maximal responsiveness from
BAGET and from our line of web services which make use of the same database.
Database Update
The genome database is updated daily at 6:00 am, GMT+1 (GMT+2 in the Summer) by a companion software which scans the NCBI
genome repository, downloads new entries and removes obsolete ones.
Upload user-provided GenBank file
BAGET accepts the submission of user-provided genomes as properly formatted GenBank files.
For obvious reasons, theses files should contain the DNA sequence. When the file is dowloaded from the NCBI,
make sure the 'GenBank (full)' option is checked in order for the DNA sequence to be appended below the annotations.
The provided genomic information is loaded and entirely processed in volatile memory (RAM) by the servers.
User data is restricted to a single user session, not shared across concurrent sessions.
All user data is removed from memory at the end of the session.
User data is therefore never saved to disk nor admin-accessible at any moment during processing to ensure maximal confidentiality.
GenBank format
The GenBank® file format is a flat text file destined to fully describe DNA or protein sequences. It consists of three parts:
• Header
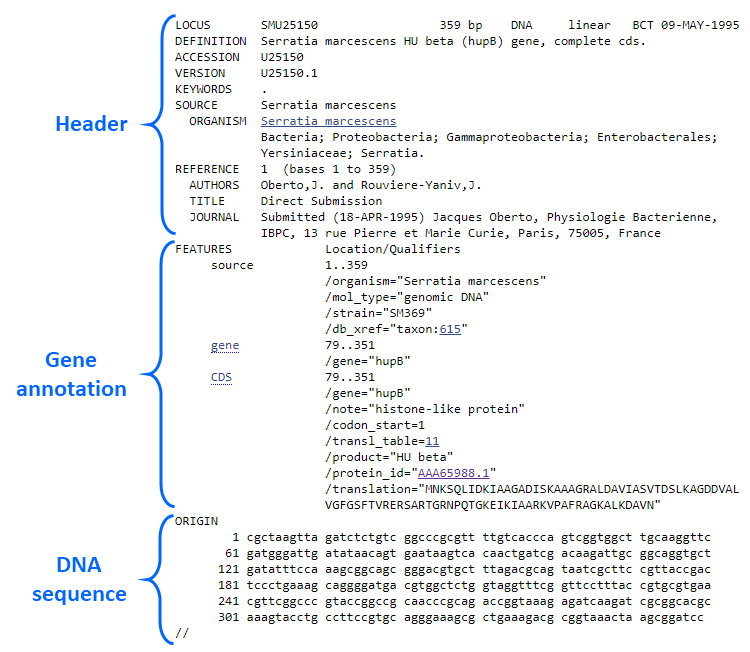
• Header
This section contains the sequence identifiers LOCUS, DEFINITION, ACCESSION/VERSION and information relevant to the source
ORGANISM. The additional fields refer to the publication record and include the AUTHORS and REFERENCE.
• Gene annotation
This section describes important characteristics of the GenBank entry as the presence of coding sequences, proteins and their definitions.
• DNA Sequence
The start of sequence section is marked by a line beginning with the word "ORIGIN" and the end of the section is marked
by a line with only "//".

Figure 1. Sample GenBank DNA file highlighting the three main sections.
Gene list
As soon as a geneome is selected by the user, the gene list is filled with all the corresponding genes lines.
Each gene line contains the following fields:
• gene start coordinate
• gene stop cordinate
• orientation
• gene definition (when known)
• gene number
• coding (CDS) or not coding (No_CDS) gene
• gene accession.version
• gene product
Each gene line can be selected independently in order to obtain the sequence context, the relative DNA sequence and corresponding protein sequence.
• gene start coordinate
• gene stop cordinate
• orientation
• gene definition (when known)
• gene number
• coding (CDS) or not coding (No_CDS) gene
• gene accession.version
• gene product
Each gene line can be selected independently in order to obtain the sequence context, the relative DNA sequence and corresponding protein sequence.
Text search
All the gene lines as displayed in the gene list are searchable by keyword or text string.
The corresponding gene lines are then listed.
Each gene line can be seletced independently in order to obtain the sequence context, the relative
DNA sequence and corresponding protein sequence.
Sequence context
The sequence context is displayed as a genetic map of ope reading frames (ORFs) centered on the selected gene in red color
always shown in its sense (5' to 3') orientation. The surrounding genes in green color are parallel to the selected gene and
those is blue color are antiparallel. The extent of the sequence context is user-selectable from 5Kb to to 30Kb in 5Kb increments.
Mouse hovering over the map reveals for each gene its id and coding function in a yellow tooltip.
All the genes populating the context map can be selected by mouse left click.
Full DNA sequence
The complete DNA sequence encompassing the selected interval depicted in the context image can be downloaded in Fasta format
for storage or further processing.
DNA sequence
The DNA sequence always displays three genes where the selected gene ORF in red color is surrounded by its upstream and
downstream gene in the same color code as the sequence context map. The intergenic reagions are indicated in default black color.
The particular cases of overlapping ORFs are taken into account and displayed in fonts with colored backgrounds.
Protein sequence
The protein sequence corresponding to the selected gene is displayed in single letter amino acid code and terminated with an asterisk.
NCBI Protein link
BAGET provides a direct external link to the NCBI repository using the accession/version of the selected gene product.
N/C refers to the non protein-coding capacity od the selected gene.
Uniprot Protein link
BAGET provides a direct external link to the Uniprot web site using the accession/version of the selected gene product.
N/C refers to the non protein-coding capacity od the selected gene.
NCBI accession/version
Every GenBank entry including full genomes, DNA and protein sequences is identified using an accession and a version number
separated by a dot. This unique identifier supersedes the previous identifier known as GI (GenInfo Identifier) number which
is now obsolete.
PDF report
BAGET results can be exported as a local file in Portable Document Format (PDF) for storage purposes or further elaboration.
BAGET provides the user a choice between the two most common "A4" and "Letter" page formats for the PDF report.
Most world countries use a page printing standard known as "A4" (297 x 210 mm) that is slightly longer and narrower
than the "U.S. letter" size (279 x 216 mm) used in North America and parts of Central and South America.
How it works
Software and Institutional Links
Archaea Software page
SynTax Prokaryotic Synteny & Taxonomy Explorer
WASPS Web Assisted Symbolic Plasmid Synteny
FITBAR Fast Inverstigation Tool for Bacterial & Archaeal Regulons
I2BC Institut de Biologie Intégrative de la Cellule
NCBI National Center for Biotechnology Information
SynTax Prokaryotic Synteny & Taxonomy Explorer
WASPS Web Assisted Symbolic Plasmid Synteny
FITBAR Fast Inverstigation Tool for Bacterial & Archaeal Regulons
I2BC Institut de Biologie Intégrative de la Cellule
NCBI National Center for Biotechnology Information